NICED makes quick detection of drug resistant H. pylori possible (GS Paper 3, Science and Technology)
Why in news?
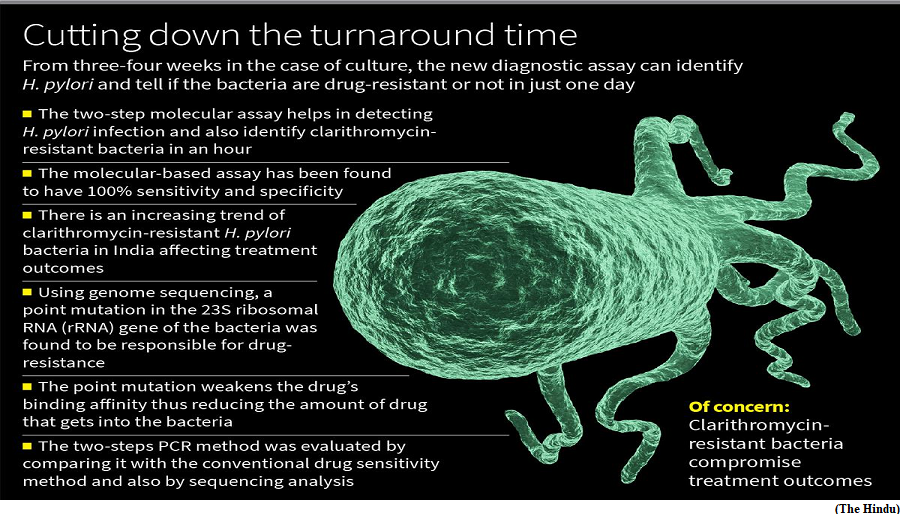
- A two-step PCR-based assay of a small region of the Helicobacter pylori (H. pylori) bacteria can help detect H. pylori infection and also identify clarithromycin-resistant bacteria and those which are drug-sensitive in six-seven hours has been developed by a team of researchers from the National Institute of Cholera and Enteric Diseases (ICMR-NICED), Kolkata.
Why it matters?
- Since H. pylori bacteria grow slowly, it takes about a week to culture the bacteria and a couple of more weeks to test for drug-sensitivity, which the new diagnostic assay bypasses. The molecular-based assay has been found to have 100% sensitivity and specificity.
- There is an increasing trend of clarithromycin-resistant H. pylori bacteria in India leading to a decreasing success rate in treating the infection.
Risk factors:
- Most of the infections caused by the bacterium H. pylori are asymptomatic, 10–15% of them develop peptic ulcer disorders or stomach cancer.
- In India, H. pylori infections affect 60-70% of the population. H. pylori infection is often acquired during childhood and remains in the stomach throughout life if not treated with antibiotics effectively.
- So, if someone suffers from gastroduodenal diseases along with the detection of H. pyloriinfection, eradication of the bacteria provides the most effective treatment.
Antibiotic resistance:
- H. pylori infection is one of the robust known risk factors for gastric cancer. As it takes three-four weeks to culture the bacteria and carry out drug-sensitivity tests, drug-resistant studies of H. pylori are seldom carried out in India.
- So, the conventionally used empirical treatment using clarithromycin is routinely used without knowing the drug-sensitivity. The growing incidence of clarithromycin-resistant bacteria is a big concern and has to be addressed as it is the most important reason for treatment failure.
How research was conducted?
- The NICED researchers undertook the study to identify the root cause of resistance toward clarithromycin and to develop a molecular-based technique for rapidly detecting antibiotic resistance.
- They turned to genome sequencing to identify that the drug resistance was due to a point mutation (A to G mutation at 2143 position) in the 23S ribosomal RNA (rRNA) gene of the bacteria.
- To confirm that the point mutation was indeed responsible for drug-resistance, the researchers isolated and amplified 617 base pairs that contained the point mutation and transferred the base pairs to drug-sensitive bacteria.
- To further confirm the role of point mutation in drug resistance, they sequenced the bacteria that had become drug-resistant after the base pairs were transferred and found that the point mutation was present in the bacteria.
PCR-based assay:
- Bioinformatics study revealed that drug-resistant and drug-sensitive strains had very different binding affinity for the drug — the drug’s binding affinity to the mutant was weaker compared with drug-sensitive bacteria.
- The researchers developed a two-step PCR-based assay to first detect H. pylori infection and then to differentiate resistant isolates from sensitive ones directly from biopsy samples.
- In the initial step of PCR, the 617 base-pair segment containing the point mutation was amplified using DNA templates isolated from biopsy samples.
- In the second PCR step, 183 base pairs amplified by the first PCR step are used as a template. For the second PCR step, two allele-specific primer sets have been designed by exploiting the point mutation in the resistant strains.
Outcome:
- The two-steps PCR method was evaluated by comparing it with the conventional drug sensitivity method and also by sequencing analysis, which showed 100% sensitivity and specificity.
Environmental factors determine height of children
(GS Paper 3, Environment)
Why in news?
- The scientists have discovered that environmental factors play a greater role than genetic variants in determining the height of children in low- and middle-income countries (LMICs) in contrast to those from European nations, where genetic aspects predominate in regulating childhood height.
- This was expounded in a study carried out by the Hyderabad-based Centre for Cellular and Molecular Biology (CSIR-CCMB) along with several other national and international institutions.

Epigenetic factors:
- While human height is strongly influenced by fixed genetic and variable environmental factors, the contribution of modifiable epigenetic factors is under-explored.
- Epigenetic factors are external influences, including lifestyle, nutrition and environment that affect the way genes work. Epigenetic changes affect gene regulation and alter gene expression but not the DNA sequence.
- Many environmental factors, including socio-economic status, nutrition and infection load are believed to influence childhood growth which plays a critical role in determining one’s height.
- The World Health Organization, 2021 estimates indicated that a large proportion of stunted children reside in LMIC, particularly in South Asia and sub-Saharan Africa where undernutrition and associated co-morbidities are more prevalent compared to high income countries (HICs).
- This offers a potential explanation for the disparity in height variation attributed to non-genetic factors between LMIC and high-income countries”.
Genome-wide association:
- Although the impact of environmental exposure during early childhood is believed to be quite significant with long-term consequences, there are no genome-wide epigenetic investigations on height in childhood especially in low and middle income countries.
- Epigenetic processes such as DNA methylation and histone modifications can influence gene expression. Methylation basically is chemical modification of DNA molecule used by cells to regulate gene expression.
- It can be influenced by environmental factors such as diet, drugs, stress, exposure to chemicals and toxins.
Highlights of the study:
- In this study, the scientists did an epigenome-wide association analysis and genome-wide association study to independently investigate links between DNA methylation and genetic variants with childhood height in five cohorts—three from India, one from Gambia and another one from the U.K.
- The scientists found a novel, robust association between methylation in the SOCS3 gene and height in children from low and middle income countries which was replicated in the HIC cohort but with a lower effect size.
- The study provides strong evidence of genome-wide DNA methylation associations with height in children from LMIC.
- The established 12,000 genetic variants were also associated with height in Indians but their effect was lower compared to the European and American counterparts.
Outcome:
- The genetic risk variations are similar for Europeans and Indians, although the magnitude differs between the two ancestries. However, the genetic risk appears to have been modified due to environmental factors in children in LMIC.
- The environmental cues that trigger the epigenetic processes in children in low and middle income countries are different in Indians and thus not influencing the epigenetic regulation of height in Europeans.



